For the x-ray examination of an object, the image contains information about the reduced intensity of
incident solar radiation inside the object. Since the absorption of x-rays is expressed in exponential
form (the Beer–Lambert law), linear information about the adsorption of radiation from the shadow
projection can be obtained using a logarithmic function. Given that a logarithm is a nonlinear
operation, even the slightest noise in the signal can result in serious errors during the reconstruction
phase. To avoid such errors, initial data averaging can be used. On the other hand, we could try to
enhance the signal noise in relation to the shadow image by optimising the exposure time to gain more
representative information. The most effective way of reducing noise in the process of reconstruction is
through a correct choice of a correction/filtering function for convolution, taking place before back
projection. In the simplest (abovementioned) case, the correction function produces two reactions of
negative adsorption around any signal or noise peak in the shadow line, but such behaviour becomes
rather risky when working with a noisy channel. This problem can be solved by choosing the convolution
function with spectral limitations (the Hamming window).
Outline of CUDA and the reconstruction process
Data reconstruction from shadow projections
The maximum size of the reconstruction cube is 1,024 × 1,024 × 1,024 voxels. We shall describe a cube of
such volume as an elementary cube. The algorithm allows
for the reconstruction of smaller cubes, but the
dimensions in all directions should be divisible by 32. In one run, the cube layer should not be more
than 128 Mb, i.e. 1/8 of an elementary cube, is reconstructed. If there are several CUDA devices
installed in the system, then the reconstruction process will be run on them simultaneously.
Shadow projections are loaded onto the RAM drive during the first run, in the amounts sufficient for the
reconstruction of the cube. For example, if the reconstruction of the cube requires 1,200 lines from the
shadow projection, but the file limit is 2,000, only the required 1,200 will be uploaded onto the RAM
and cached. Since it is very likely that the segments of projection will overlap in the process of the
next cube reconstruction, we can use the data, which has already been uploaded, one more time, only then
having to upload the new lines.
Connecting to the server and task distribution
Broadcasting of UDP packets is used to find clients and servers. The server sends out a notification upon
start-up and subsequently sends notifications at five-minute intervals. The client, when launched, sends
out a broadcast query which, in turn, sends out an immediate server response command. The client, having
received a notification about an available server, connects to it via TCP, which is used to send
executive commands and tasks and receive reports about task termination. All clients, servers and tasks
have unique identifiers.
When the connection to one of the servers is lost, the server is considered to have stopped, and its
tasks are distributed among the others. When a new server is connected, the tasks are neither taken away
from other servers nor redirected towards it. That is why, to provide an even load, it is very important
to find all servers before the start of reconstruction task distribution. As more cubes are being made
ready, it becomes possible to launch integration tasks. These tasks are directed to a random server. The
tasks of reconstruction and integration are performed by servers simultaneously.
Tasks may come from several clients. The server processes them in order of their appearance (FIFO). The
client operates best when sending out reconstruction tasks from one layer of elementary cubes to one
server. This is to reduce the amount of data necessary for the server to begin reconstruction. The
amount of server RAM determines the strategy of reconstruction and compression of cubes. If there is a
large amount of memory, (more than 20 GB) then the compression of the previous cube and the
reconstruction of the next one is done simultaneously, otherwise the reconstruction of the next cube
will not start until the previous one has been fully processed, i.e. compressed and saved to the hard
drive.
The main stages of reconstruction
First of all, projection segments, into which the reconstructed volume is being projected, are determined
after accounting for perspective. The projections needed for reconstruction are identified on the basis
of this data. If the RAM cache contains segments of files that are not on this list, they will be
deleted from the memory.
The reconstruction thread addresses the cache for a segment of the projection, the memory unit containing
the projection (considering the page size of 4,096 bytes) has its memory locked, which prevents it from
being replaced, and the pointer is passed along to the CUDA procedure. All of the following operations
are held in the memory of the accelerator. First of all, the 12-bit storage format is decompressed into
a float data type, and all further calculations are conducted in float. During decompression, the
segment is trimmed on both the left and right to fit the length of the reconstructed block, with some
allowance for convolution when filtering the projection. Some preliminary processing is conducted:
inversion, gamma-correction, filtering by averaged projection, and conducting the necessary number of
smoothing iterations. Then, the convolution is carried out and the segment again is trimmed on the left
and on the right to the minimum width needed for back projection. In this way, the projections are
processed until there is no free memory on the video card. Thus, an array of segments ready for back
projection is formed.
The essence of back projection lies in summing the weights of all the x-ray projections of a particular
point for every voxel. One CUDA-core request manages calculations for 32 voxels placed one above the
other. At the same time, a cycle is made through all projection segments uploaded to the memory. This
allows for the storage of all intermediate data in registers without any intermediary storage. At the
end of the calculation stage, the value of the voxel's density is recorded in the memory. The
back-projection kernel has two possible implementations. One of them, the slow implementation, includes
checks to see if there is a point on the projection to draw data from, since such a point is not
necessarily present. The fast implementation does not account for checks. The appropriate implementation
is chosen automatically. The selection of a value from the projection is carried out according to the
‘nearest neighbour algorithm’. The use of texture memory, during the early stages, led to a sharp
reduction in efficiency. At this stage, when projection arrays were used, it appeared impossible to
create texture arrays due to the limitations posed by CUDA 4.2.
If there isn't enough memory, the calculations might need several iterations of projection uploads and
back projections. When the calculations are completed, the obtained voxels are evaluated. Maximum and
minimum density values are calculated and a bar chart is created. Plotting the bar chart and copying the
results onto the RAM drive are done simultaneously.
When there are several CUDA devices, the segments of a reconstructed cube are processed simultaneously.
Once the reconstruction of a cube has completed, another thread takes on the task of compressing the
cube, and the reconstruction thread immediately undertakes the next task, providing that there is enough
RAM capacity. Dedicating and freeing up large segments of memory proved to be a very time-consuming
operation, which is why all large already-obtained memory units have been used in subsequent
calculations. This is why, some CUDA memory is dedicated to voxels, some for a series of intermediary
buffers, and the rest of the memory is allocated to projections. The projection stores are managed
manually.
Compression
Compression to octree is conducted in several threads. The number of threads depends on the number of
processor cores. Every thread processes its own octant. It is then saved in a file indicated by the
task's reconstruction client. A notification is finally sent that the processing of the cube is
complete.
Client
The client maintains a database of cubes in the form of an SQLite
file. Cube characteristics, reconstruction time and other values are stored there. Upon finishing the
reconstruction, the client program closes, and package tasks for reconstruction can be organised.
A sample CUDA code
The core (measureModel.cu) operates the first run while plotting the bar chart. The file is given to
demonstrate the general form of CUDA code in an actual working project.
struct MinAndMaxValue
{
float minValue;
float maxValue;
};
__global__ void
cudaMeasureCubeKernel(float* src, MinAndMaxValue* dst, int ySize, int sliceSize, int blocksPerGridX)
{
int x = blockDim.x * blockIdx.x + threadIdx.x;
int z = blockDim.y * blockIdx.y + threadIdx.y;
int indx = z * blocksPerGridX * blockDim.x + x;
MinAndMaxValue v;
v.minValue = src[indx];
v.maxValue = v.minValue;
for (int i = 0; i < ySize; ++i)
{
float val = src[indx];
indx += sliceSize;
if (v.minValue > val)
v.minValue = val;
if (v.maxValue < val)
v.maxValue = val;
}
__shared__ MinAndMaxValue tmp[16][16];
tmp[threadIdx.y][threadIdx.x] = v;
__syncthreads();
if (threadIdx.y == 0)
{
for (int i = 0; i < blockDim.y; i++)
{
if (v.minValue > tmp[i][threadIdx.x].minValue)
v.minValue = tmp[i][threadIdx.x].minValue;
if (v.maxValue < tmp[i][threadIdx.x].maxValue)
v.maxValue = tmp[i][threadIdx.x].maxValue;
}
tmp[0][threadIdx.x] = v;
}
__syncthreads();
if (threadIdx.x == 0 && threadIdx.y == 0)
{
for (int i = 0; i < blockDim.y; i++)
{
if (v.minValue > tmp[0][i].minValue)
v.minValue = tmp[0][i].minValue;
if (v.maxValue < tmp[0][i].maxValue)
v.maxValue = tmp[0][i].maxValue;
}
dst[blockIdx.y * blocksPerGridX + blockIdx.x] = v;
}
}
void Xrmt::Cuda::CudaMeasureCube(MeasureCubeParams params)
{
int threadsPerBlockX = 16;
int threadsPerBlockZ = 16;
int blocksPerGridX = (params.xSize + threadsPerBlockX - 1) / threadsPerBlockX;
int blocksPerGridZ = (params.zSize + threadsPerBlockZ - 1) / threadsPerBlockZ;
int blockCount = blocksPerGridX * blocksPerGridZ;
dim3 threadDim(threadsPerBlockX, threadsPerBlockZ);
dim3 gridDim(blocksPerGridX, blocksPerGridZ);
MinAndMaxValue *d_result = nullptr;
size_t size = blockCount * sizeof(MinAndMaxValue);
checkCudaErrors(cudaMalloc( (void**) &d_result, size));
cudaMeasureCubeKernel<<<gridDim, threadDim>>>(
params.d_src,
d_result,
params.ySize,
params.xSize * params.zSize,
blocksPerGridX
);
checkCudaErrors(cudaDeviceSynchronize());
MinAndMaxValue *result = new MinAndMaxValue[blockCount];
checkCudaErrors(cudaMemcpy(result, d_result, size, cudaMemcpyDeviceToHost));
params.maxValue = result[0].maxValue;
params.minValue = result[0].minValue;
for (int i = 1; i < blockCount; i++)
{
params.maxValue = std::max(params.maxValue, result[i].maxValue);
params.minValue = std::min(params.minValue, result[i].minValue);
}
checkCudaErrors(cudaFree(d_result));
}
__global__ void
cudaCropDensityKernel(float* src, int xSize, int ySize, int sliceSize, float minDensity, float maxDensity)
{
int x = blockDim.x * blockIdx.x + threadIdx.x;
int z = blockDim.y * blockIdx.y + threadIdx.y;
int indx = z * xSize + x;
for (int i = 0; i < ySize; ++i)
{
float val = src[indx];
if (val < minDensity)
src[indx] = 0;
else if (val > maxDensity)
src[indx] = maxDensity;
indx += sliceSize;
}
}
void Xrmt::Cuda::CudaPostProcessCube(PostProcessCubeParams params)
{
int threadsPerBlockX = 16;
int threadsPerBlockZ = 16;
int blocksPerGridX = (params.xSize + threadsPerBlockX - 1) / threadsPerBlockX;
int blocksPerGridZ = (params.zSize + threadsPerBlockZ - 1) / threadsPerBlockZ;
dim3 threadDim(threadsPerBlockX, threadsPerBlockZ);
dim3 gridDim(blocksPerGridX, blocksPerGridZ);
cudaCropDensityKernel<<<gridDim, threadDim>>>(
params.d_src,
params.xSize,
params.ySize,
params.xSize * params.zSize,
params.minDensity,
params.maxDensity
);
checkCudaErrors(cudaDeviceSynchronize());
}
__global__ void
cudaBuildGistogramKernel(
float *src,
int count,
int *d_values,
int minValue,
int barCount
)
{
int barFrom = threadIdx.x * ValuesPerThread;
int blockIndx = SplitCount * blockIdx.x + threadIdx.y;
int blockLen = count / (SplitCount * SplitCount2);
unsigned int values[ValuesPerThread];
for (int i = 0; i < ValuesPerThread; i++)
{
values[i] = 0;
}
src += blockLen * blockIndx;
for(int i = blockLen - 1; i >= 0; i--)
{
float v = src[i];
int indx = (int)((v - (float)minValue) * DivideCount) - barFrom;
if (indx >= 0 && indx < ValuesPerThread)
values[indx]++;
}
for (int i = 0; i < ValuesPerThread; i++)
{
d_values[blockIndx * barCount + barFrom + i] = values[i];
}
}
__global__ void
cudaSumGistogramKernel(
int *d_values,
int barCount
)
{
int barIndx = blockIdx.x * ValuesPerThread + threadIdx.x;
unsigned int sum = 0;
d_values += barIndx;
for (int i = 0 ; i < SplitCount * SplitCount2; i++)
{
sum += d_values[i * barCount];
}
d_values[0] = sum;
}
void Xrmt::Cuda::CudaBuildGistogram(BuildGistogramParams params)
{
checkCudaErrors(cudaMemcpyAsync(
params.h_dst, params.d_src, params.count * sizeof(float), cudaMemcpyDeviceToHost, 0
));
int minValue = (int)(params.minDensity - 1);
minValue = std::max(minValue, -200);
int maxValue = (int)params.maxDensity;
maxValue = std::min(maxValue, 200);
int barCount = (maxValue - minValue + 1) * DivideCount;
int x32 = barCount % ValuesPerThread;
barCount += (x32 == 0) ? 0 : ValuesPerThread - x32;
dim3 threadDim(barCount / 32, SplitCount);
dim3 gridDim(SplitCount2);
int *d_values = nullptr;
size_t blockValuesCount = SplitCount2 * SplitCount * barCount;
size_t d_size = blockValuesCount * sizeof(int);
checkCudaErrors(cudaMalloc( (void**) &d_values, d_size));
cudaBuildGistogramKernel<<<gridDim, threadDim>>>(
params.d_src,
params.count,
d_values,
minValue,
barCount
);
cudaSumGistogramKernel<<<dim3(barCount / ValuesPerThread), dim3(ValuesPerThread)>>>(
d_values,
barCount
);
checkCudaErrors(cudaDeviceSynchronize());
params.histogramValues.resize(barCount);
checkCudaErrors(cudaMemcpy(
params.histogramValues[0],
d_values,
barCount * sizeof(int),
cudaMemcpyDeviceToHost
));
checkCudaErrors(cudaFree(d_values));
params.histogramStep = 1.0 / DivideCount;
params.histogramStart = (float)minValue;
}
Question #2. The camera that you used (PIXIS-XF, I think) produces 2,048 × 2,048 images.
However, in the article you mentioned ‘up to 8,000 × 8,000’. How did you reach this? Are you supposed
to move the object, or move the camera vertically and horizontally and later patch the
images?
Yes, you are right. The camera used in the microtomograph is the PIXIS-XF: 2048B. The size of the
reconstructed image is, according to the requirements, 8,000 × 8,000 × 20,000. 8,000 in width and depth.
This means that the camera sees only a part of the image, but the algorithm allows for greater
reconstruction by way of many circular projections. Indeed, this accounts for a decrease in quality of
the reconstruction along the edges. As for the height, we simply move the table and create a new
reconstruction; the results are then patched.
Question #3. Are all the demo images, which are given in the video at the end of the
article, received from 360-degree projections? If so, then it's rather good; 360-degree projections
in increments of 1 degree are very rare; usually one has to progress in increments of a third or
quarter of a degree, otherwise the signs of reconstruction will be visible. I think there was a
formula for an optimal number of projections for a specified resolution, but I can't remember it
exactly.
In our case, 180–360 projections were used. Various algorithms may provide different reconstruction
quality depending on the number of projections, but the rule of thumb is that while the number of
projects increase, the quality increases too. For our task, it was sufficient to use 180–360 projections
to provide high quality.
The algorithm reacts well to skipped frames at the cost of a slight reduction of resolution in the
direction where the gaps are. Besides, the camera had a tendency to get overheated and its ‘brightness’
wavered, which is why some frames were discarded as defective. The others were normalised according to
average brightness.
Question #4. I didn't quite understand about the speed of the camera. The specification
says that it's dual-frequency 100 kHz or 2 MHz. If it's a four-megapixel camera, does it mean that it
shoots a frame every two seconds at the maximum frequency?
Yes, you are correct. According to the camera
documentation, its specifications are the following: ADC speed/bits 100 kHz/16-bit and 2 MHz/16-bit.
That means that if one dot of the image has a 16-bit capacity, we will get a frequency for the whole
image of 2,000,000 / (2,048 * 2,048) = 0.47 Hz.
Question #5. Do you move the manipulator step by step or continuously? How long
does it take to
scan the typical samples that were shown in the video?
The manipulator (a table) moves in steps. We turned it around a corner, stopped, made a shot, turned it
again etc. The average scanning time was 11 minutes for 378 frames.
Question #6. I'd be interested to learn about the 12-bit setup. It stands to reason that
one can cut off four bits and pack each two-pixel unit into three bytes for storage and
transferring. But for the reconstruction you will have to expand each pixel into a minimum of two
bytes... Or is all of your mathematics based on 12-bit? In that case, how did you solve the problem of
the alignment of 1.5-byte pixels in storage?
Here is the file structure for the 12-bit side projections.
- Pixels are packed bit-by-bit and placed in lines, so that two points take up three
bytes;
it is being understood that the width of the projection image is divisible by two
according
to
the formula: lineWidthInBytes = width() * 3 / 2.
- Usually the whole range of data is not loaded at once, but ‘windows’ are used where only the
necessary portion of the image is loaded while the left and right borders are supposed to be aligned
so as to be divisible by four pixels.
- After the ‘window’ is loaded further, work is conducted via the function of complete
import/export of this window into float or quint16 arrays. During the import/export process, the
data is converted from 12-bit to the necessary number of bits.
- In our case, the benefit from shrinking the size of projection files was greater than the loss of
efficiency caused by it. Besides, the entire files were not loaded, instead, the ‘windows’ were,
intended to be processed on a particular cluster engine. Indeed, losses in productivity were even
smaller thanks to server multitasking.
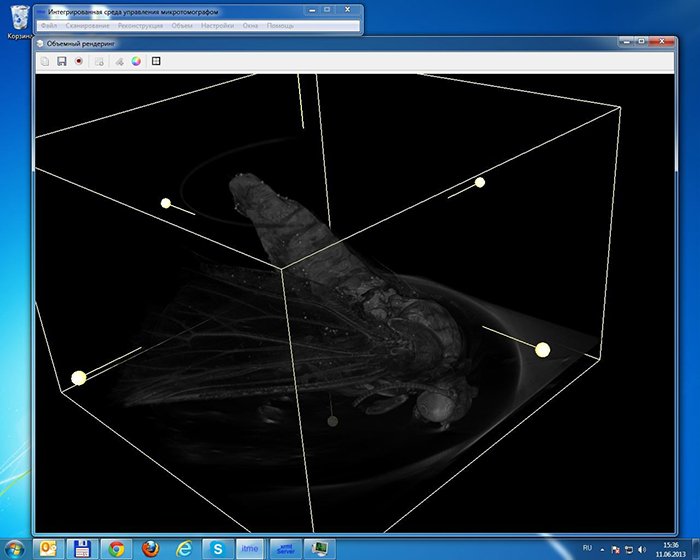
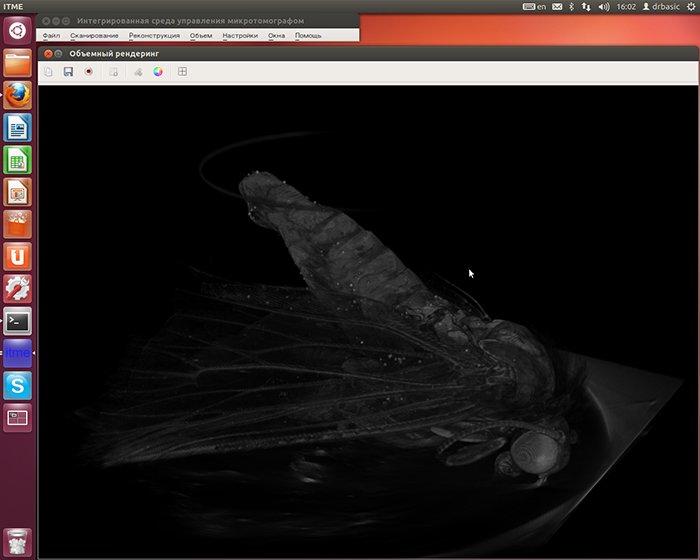
Volume (voxel) rendering in real time
Another interesting task was rendering the model with the use of voxels in real time. Apart from voxels,
the model itself had to render instruments such as the minimum bounding box and sectional plane in 3D,
hide half of the object along the sectional plane as well as hide the object outside the box. The
sectional plane has an element of user interface in the form of a ‘switch’ (which at the
same time
corresponds to the normal vector) allowing for the window of the projection slice to be moved along the
section plane and for the plane itself to move along the normal vector. The window of the projection
slice can also be stretched beyond its boundaries and rotated. The bounding box displays both its edges
and the same kinds of ‘switches’ as on the plane, which also allow its size to be changed. All these
manipulations are conducted in real time within the volume space of the model.
A sufficient frame rate was sustained during the operation by means of a variable level of granularity,
as the data is uploaded automatically once the bounding box is shrunk.
Special sets of mathematical functions were implemented to manipulate interface elements in 3D. The
initial creation of the section plane, if described in general terms without any formulae, looks like
this: a user double-clicks on any point of the model, upon which the point is fixed. Upon further
movement of the mouse, it serves as the starting point to display the normal vector to the created
plane. However, the plane has already been created in practical terms, lying on the half-line starting
from the display coordinates of the mouse cursor and pointing into the depth of field, non-parallel with
the perspective lines so that the half-line is projected onto the screen as a point and the plane
orientation changes dynamically with movement of the mouse; the user sees it instantly on screen. To
finish the creation, the mouse should be clicked one more time to lock the plane's orientation.
In addition to the elements of model volume manipulation, there are instruments for managing colour
spectra of densities and transparency levels.
The self-rendering voxel, the slicing by the section plane and the bounding box were implemented with the
help of the VTK (Visualization Toolkit) library functionality.
A list of mathematical functions
- Plane
functions:
- Mathematical functions for geometrical calculations:
- Mathematical functions used during reconstruction:








 2002–2026
2002–2026